Section: New Results
Head Tracking and Flagellum Tracing for Sperm Motility Analysis
Participants : Huei Fang Yang, Xavier Descombes, Grégoire Malandain, Sylvain Prigent.
This work is supported in part by ANR MOTIMO project.
Sperm quality assessment plays an important role in human fertility and animal breeding. One of the most important attributes for evaluating semen quality is sperm motility, according to the World Health Organization (WHO) report. When performed manually, semen analysis based on sperm motility is labor-intensive and subject to intra- and inter-observer variability. Computer-assisted sperm analysis (CASA) systems, in contrast, provide rapid and objective semen fertility assessment. In addition, they also offer a means of statistical analysis that may not be achieved by visual assessment. Hence, automated sperm motility analysis systems are highly desirable.
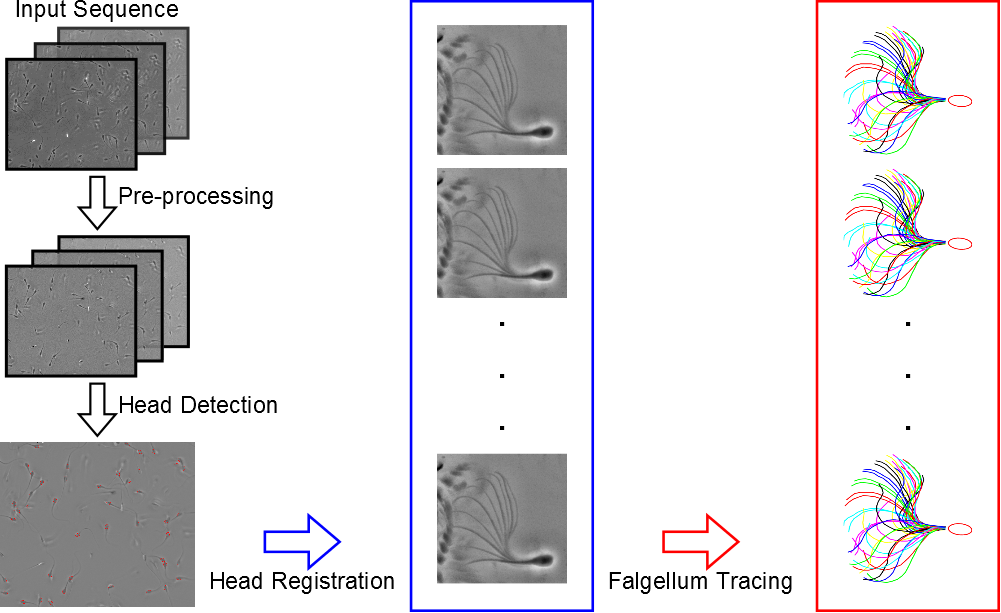
We present a computational framework designed to track the heads and trace the tails for quantitative analysis of sperm motility, which is illustrated in Figure 3 . Our framework includes 3 modules: head detection, head registration, and flagellum tracing. These modules are performed sequentially to obtain the head trajectories and flagellar beat patterns. First, the head detection module detects the sperm heads in the first image of the image data using a Multiple Birth and Cut (MBC) algorithm. The detections are the inputs to the head registration module for obtaining the head trajectories and angles of head rotation. We use a block matching method to register the heads in the subsequent images with respect to the positions and angles of those detected in the first image. This is different from other tracking methods that consider only the head positions. Finally, we propose a flagellum tracing algorithm, based on a Markov chain Monte Carlo (MCMC) sampling method, to obtain the flagellar beat patterns.
We validate our framework using two microscopy image sequences of ram semen samples that were imaged at two different conditions, at which the sperms behave differently. The results show the effectiveness of our framework [19] .
|